Our lab is interested in understanding the structure, dynamics, and functions of the signaling network involving receptor tyrosine kinases (RTKs) and their downstream effectors such Ras, PI3K and MAPK/ERK pathways. We use an interdisciplinary approach including fluorescent live cell imaging, genetics, synthetic biology, optogenetics, and mathematical modeling to uncover the mechanisms and properties of the network in cell biology, as well as in vivo models and patient samples to investigate the implications in cancer treatment.
Research Highlights
A method for massively multiplexed biosensor tracking in barcoded cells
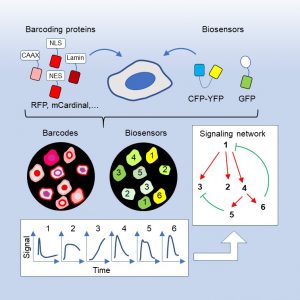
Genetically encoded fluorescent biosensors are powerful tools for real-time monitoring of biochemical activities in cells, but their multiplexing capacity is severely limited by the available spectral space. We overcome this problem by developing a set of barcoding proteins that are spectrally separable from commonly used FRET (fluorescence resonance energy transfer)-based and single-fluorophore biosensors. Combinations of barcoding proteins targeted to different subcellular locations can generate over 100 barcodes. Mixed populations of barcoded cells expressing different biosensors are spectrally imaged and computationally unmixed to achieve massively multiplexed tracking of biochemical activities in live cells. Moreover, we develop deep learning models to automate barcode reading in various cell lines. Importantly, different biosensors in the cell mixture show highly coordinated activities, thus facilitating the delineation of their temporal relationship. By simultaneously tracking multiple biosensors in the receptor tyrosine kinase (RTK) signaling network, we provided insights into the distinct mechanisms of effector adaptation, cell autonomous and non-autonomous effects of KRAS mutations, as well as the complex interactions between different nodes in the network. Together, these results demonstrate that biosensor barcoding presents a powerful and easily scalable method for deciphering the complexity of signaling networks as well as their interactions between cells.
References
- Yang JM†, Chi WY, Liang J, Takayanagi S, Iglesias P, Huang CH†. Deciphering cell signaling networks with massively multiplexed biosensor barcoding. Cell. 2021; 184(25), 6193–6206. PMID: 34838160. [pdf] (Cover Article)
- Chi WY, Au G, Liang J, Chen CC, Huang CH†, Yang JM†. Imaging and analysis for simultaneous tracking of fluorescent biosensors in barcoded cells. STAR Protocols. 2022; 3(3), 101611. [pdf]
- Wang S*, Chi WY*, Au G, Huang CC, Yang JM†, Huang CH†. Reconstructing signaling networks using biosensor barcoding. Methods Mol Biol. 2024;2800:189–202. [pdf]
Integration of chemical and mechanical stimuli by Ras/PI3K/ERK signaling
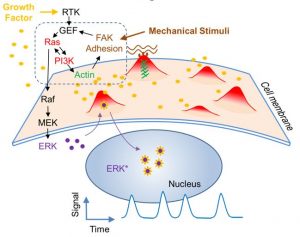
The Ras/PI3K/ERK signaling pathway regulates diverse cellular processes in response to environmental stimuli and contains important therapeutic targets for cancer. Recent single cell studies revealed stochastic pulses of ERK activation, the frequency of which determines functional outcomes such as cell proliferation. Using fluorescent live cell imaging including total internal reflection and lattice light sheet microscopy, we found that ERK pulses were initiated by localized protrusive activities. Chemically and optogenetically induced protrusions triggered ERK activation through various entry points into the feedback loop involving Ras, PI3K, the cytoskeleton, and cellular adhesion. The excitability of the protrusive signaling network drove stochastic ERK activation in unstimulated cells and oscillations upon growth factor stimulation. Importantly, protrusions allowed cells to sense combined signals from substrate stiffness and the growth factor. Thus, by uncovering the basis of ERK pulse generation we demonstrate how signals involved in cell growth and differentiation are regulated by dynamic protrusions that integrate chemical and mechanical inputs from the environment.
References
- Yang JM, Bhattacharya S, West-Foyle H, Hung CH, Wu TC, Iglesias PA & Huang CH. “Integrating chemical and mechanical signals through dynamic coupling between cellular protrusions and pulsed ERK activation.” Nat Commun. 2018; 9(1), 4673. PMID: 30405112. [pdf]
Ras-PI3K signaling in cell migration
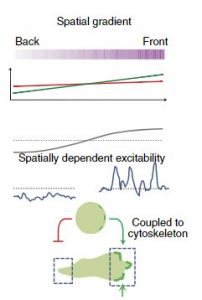 Cell migration is a fundamental cellular phenomenon that plays important roles in normal physiology, such as embryonic development, tissue regeneration, wound healing, and immune responses. Abnormal cell migration underlies the pathogenesis of diseases such as asthma, inflammatory diseases, and metastatic cancers. Cell migration requires the generation of protrusions at the leading edges. The protrusions are associated with the activation of signal transduction proteins such as Ras GTPases and PI3Ks, as well as regulators of actin polymerization such as the SCAR/WAVE complex. It was not clear how these events were triggered in the absence of stimuli or what roles they played in the generation of protrusions. To answer these questions, I analyzed the spatiotemporal patterns of biosensors for signal transduction and cytoskeletal events in the model organism Dictyostelium and found that they belong to two separable networks with distinct kinetics and functions. The cytoskeletal network, comprising SCAR/WAVE, Arp2/3, F-actin, and coronin, displays oscillations with a period of 10 sec that only causes small-amplitude cell boundary undulations. In contrast, the signal transduction network, involving Ras GTPases, PI3Ks, and Rac GTPases, displays features of excitability such as wave propagation, refractoriness, and all-or-none responses, even in the absence of cytoskeletal turnover. These results suggest that the cytoskeletal network functions as an “idling” motor force that by itself does not make cells move. Instead, the intermittent, more prolonged “pacemaker” activity of the signaling network is required to generate protrusions. The excitability of signaling networks explains how it can be triggered stochastically by noise to mediate random movement. Moreover, we found that the threshold of the excitable signaling network is modulated by an upstream gradient sensing module mediated by a local excitation, global inhibition (LEGI) mechanism, allowing cells to respond only to the spatial difference in chemoattractant concentration. The salient features of the coupling between adaptive and excitable networks were found in Dictyostelium responding to the chemoattractants cAMP and folic acid as well as human neutrophils responding to fMLP, suggesting an evolutionarily conserved mechanism for eukaryotic chemotaxis. Based on this scheme we were able to build mathematical models that quantitatively describe the responses of cells to chemoattractant stimuli. Taken together, our work shows the elegant strategy cells employ for directed migration.
Cell migration is a fundamental cellular phenomenon that plays important roles in normal physiology, such as embryonic development, tissue regeneration, wound healing, and immune responses. Abnormal cell migration underlies the pathogenesis of diseases such as asthma, inflammatory diseases, and metastatic cancers. Cell migration requires the generation of protrusions at the leading edges. The protrusions are associated with the activation of signal transduction proteins such as Ras GTPases and PI3Ks, as well as regulators of actin polymerization such as the SCAR/WAVE complex. It was not clear how these events were triggered in the absence of stimuli or what roles they played in the generation of protrusions. To answer these questions, I analyzed the spatiotemporal patterns of biosensors for signal transduction and cytoskeletal events in the model organism Dictyostelium and found that they belong to two separable networks with distinct kinetics and functions. The cytoskeletal network, comprising SCAR/WAVE, Arp2/3, F-actin, and coronin, displays oscillations with a period of 10 sec that only causes small-amplitude cell boundary undulations. In contrast, the signal transduction network, involving Ras GTPases, PI3Ks, and Rac GTPases, displays features of excitability such as wave propagation, refractoriness, and all-or-none responses, even in the absence of cytoskeletal turnover. These results suggest that the cytoskeletal network functions as an “idling” motor force that by itself does not make cells move. Instead, the intermittent, more prolonged “pacemaker” activity of the signaling network is required to generate protrusions. The excitability of signaling networks explains how it can be triggered stochastically by noise to mediate random movement. Moreover, we found that the threshold of the excitable signaling network is modulated by an upstream gradient sensing module mediated by a local excitation, global inhibition (LEGI) mechanism, allowing cells to respond only to the spatial difference in chemoattractant concentration. The salient features of the coupling between adaptive and excitable networks were found in Dictyostelium responding to the chemoattractants cAMP and folic acid as well as human neutrophils responding to fMLP, suggesting an evolutionarily conserved mechanism for eukaryotic chemotaxis. Based on this scheme we were able to build mathematical models that quantitatively describe the responses of cells to chemoattractant stimuli. Taken together, our work shows the elegant strategy cells employ for directed migration.
References
- Huang CH, Tang M, Shi C, Iglesias PA and Devreotes PN. “An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration.” Nat Cell Biol. 2013; 15(11): 1307-1316. [pdf]
- Tang M, Wang M, Shi C, Iglesias PA and Devreotes PN, Huang CH. “Evolutionarily conserved coupling of adaptive and excitable networks mediates eukaryotic chemotaxis.” Nat Commun. 2014; 5: 5175. [pdf]
- Zhan H, Bhattacharya S, Cai H, Iglesias PA, Huang CH†, & Devreotes PN†. An Excitable Ras/PI3K/ERK Signaling Network Controls Migration and Oncogenic Transformation in Epithelial Cells. Developmental Cell. 2020; 54(5), 608–623.e5. [pdf]
