The assessment of protective immunity conferred by vaccination has three essential steps. First, an immune response is elicited by immunization with a candidate vaccine. Second, the immunized mice, along with unimmunized controls, are challenged with antigen-expressing tumors. Finally, the antitumor effects in the immunized mice and unimmunized controlsis are compared.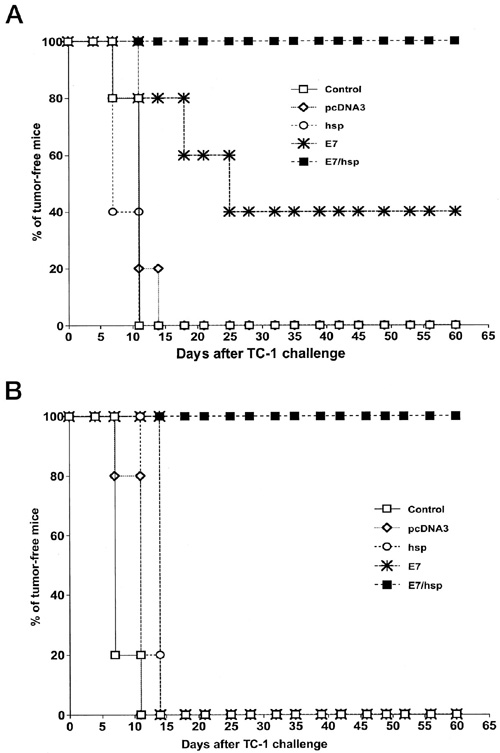
Vaccination with E7-HSP70 DNA protects mice against the growth of TC-1 tumors.C57BL/6 mice were immunized with empty plasmid (pcDNA3), HSP70 DNA (HSP), E7 DNA (E7), or E7-HSP70 DNA (E7/HSP) via a gene gun. One week after the last vaccination, mice were challenged with 5 x 104 TC-1 cells/mouse s.c. in the right leg. The mice were monitored for evidence of tumor growth by palpation and inspection twice a week. A, each mouse received 2 µg of DNA vaccine. One week later, the mice were revaccinated with the same type and amount of DNA as the first vaccination. B, each mouse received only one injection of 2 µg of DNA vaccine without a further booster. (Enhancement of DNA Vaccine Potency by Linkage of Antigen Gene to an HSP70 Gene)
To assess the therapeutic effects of vaccination, one first challenges mice with antigen-expressing tumors. Second, some mice are immunized with a candidate vaccine while unimmunized mice are used as controls. Finally, the antitumor effect in immunized subjects and unimmunized controls is evaluated and compared.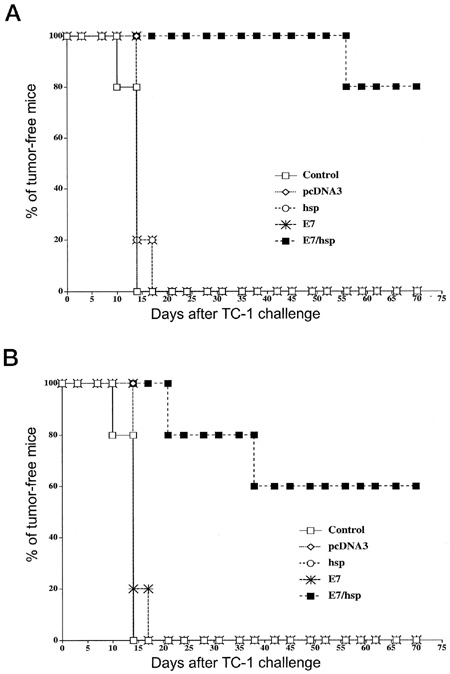
Vaccination with E7-HSP70 DNA eradicates pre-existing TC-1 tumor cells. Each mouse was initially challenged with 2 x 104 TC-1 cells s.c., followed by DNA vaccination with empty plasmid (pcDNA3), HSP70 DNA (HSP), E7 DNA (E7), or E7-HSP70 DNA (E7/HSP) via a gene gun. The mice were monitored for evidence of tumor growth by palpation and inspection twice a week. A, each mouse received 2 µg of DNA vaccine 3 days and 10 days after the tumor challenge. B, each mouse received 2 µg of DNA vaccine only 3 days after the tumor challenge. No further booster was given. (Enhancement of DNA Vaccine Potency by Linkage of Antigen Gene to an HSP70 Gene)
The Lung Metastasis Model to Assess the Therapeutic Effect of a Cancer Vaccine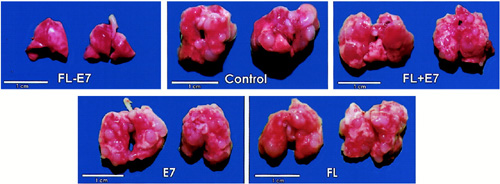
Representative gross pictures of the lung tumors in each vaccinated group. After in vivo tumor treatment experiments against preexisting metastatic TC-1 tumor cells, there are multiple grossly visible lung tumors in unvaccinated control mice and mice vaccinated with FL, wild-type E7 DNA, or FL mixed with E7 DNA. The lung tumors in the FL-E7 vaccinated group cannot be seen at the magnification provided in this figure. (Enhancement of DNA Vaccine Potency by Linkage of Antigen Gene to a Gene Encoding the Extracellular Domain of Fms-like Tyrosine Kinase 3-Ligand)
Cell culture involves the maintenance and growth of the cells of multicellular organisms outside the body in specially designed containers and under precise conditions of temperature, humidity, nutrition, and freedom from contamination. In a broad sense, cells, tissues, and organs that are isolated and maintained in the laboratory are considered the objects of tissue culture. Scientists to use cell cultures for experimental studies and for biological assays.
The cytotoxic T lymphocyte assay is used to detect the cytolytic activity of antigen-specific T cells. Target cells are labeled with radioactive sodium chromate, lactate dehydrogenase (LDH), or other readily detectable compound. A sample containing T cells is then added to the mixture. When these target cells are killed, the chromate or LDH is is released, providing a measure of cytotoxicity.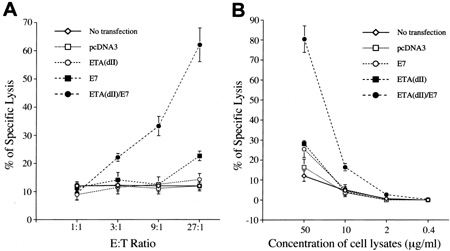
CTL assays. A, CTL assays to demonstrate enhanced presentation of E7 through the MHC class I pathway of cells transfected with ETA(dII)/E7 DNA. 293 Db, Kb cells transfected with various DNA constructs served as target cells. These CTL assays were performed with various E:T ratios using Db-restricted E7-specific CD8+ T cells as target cells. B, CTL assays to demonstrate enhanced MHC class I presentation of E7 in bone marrow-derived DCs pulsed with cell lysates containing chimeric ETA(dII)/E7 protein. Bone marrow-derived DCs were pulsed with cell lysates from various DNA-transfected 293 Db, Kb cells at different concentrations as described in “Materials and Methods.” These assays were performed at a fixed E:T (9:1) ratio using Db-restricted E7-specific CD8+ T cells as effector cells. (Cancer Immunotherapy Using a DNA Vaccine Encoding the Translocation Domain of a Bacterial Toxin Linked to a Tumor Antigen)
A procedure in which proteins or DNA are separated on a gel for their detection, characterization, or quantification. Sodium dodecyl sulfate – polyacrylamide gel electrophoresis (SDS-PAGE) is used for protein, while agarose gel electrophoresis is used for DNA. Electrophoresis separates the components of the sample into discrete zones separated by their size (or alternatively, by charge), with each zone containing one or more proteins or DNA. Sometimes, gel electrophoresis is followed by a western blot anaysis.
Enzyme-linked immunosorbent assay (ELISA) is used to detect the presence of antibodies or antigen in a sample. In a direct ELISA, antigen is coated on the surface of plastic wells. Serum containing antibodies against this antigen are added to the wells. Labeled anti-Ig antibody is added to the wells to bind with unlabeled antibody, followed by a reaction with the appropriate substrate that changes the color of the product. This assay can also be modified for the detection of cytokines or other proteins.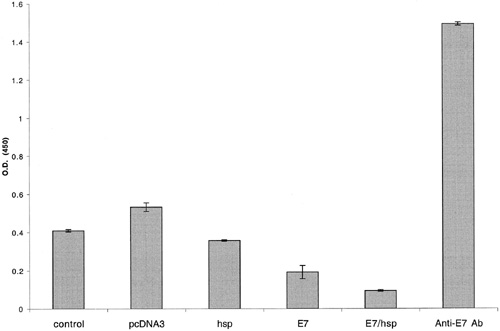
E7-specific antibody responses in C57BL/6 mice immunized with various recombinant DNA vaccines. C57BL/6 mice were immunized with control plasmid (no insert), wild-type HSP70, wild-type E7, or E7-HSP70 DNA via a gene gun. Serum samples were obtained from immunized mice 14 days after vaccination. The presence of the E7-specific antibody was detected by ELISA using serial dilution of sera. The results from the 1:80 dilution are presented showing the mean absorbance (A450 nm) SE. (Enhancement of DNA Vaccine Potency by Linkage of Antigen Gene to an HSP70 Gene)
The enzyme-linked immunospot (ELISPOT) assay is designed to enumerate cytokine producing cells in a single cell suspension. This method has the advantage of requiring a minimum of in vitro manipulations allowing cytokine production analysis as close as possible to in vivo conditions. This technique is designed to determine the frequency of cytokine producing cells under a given stimulation, and the follow-up of such frequency during a treatment and/or pathological state. To perform the ELISPOT assay, cytokine-specific antibodies are first attached to the surface of a plastic well. Next, cytokine-secreting cells (i.e. activated T cells) are added to the wells. Cytokine that is secreted by these cells are bound by cytokine-specific antibodies. Finally, cytokine-secreting cells are revealed by a second cytokine-specific antibody that is enzyme-labeled; after a reaction with the appropriate substrate, each cell secreting cytokine is revealed as a spot.
For more detailed information about the ELISPOT assay and how it is performed, please refer to this .pdf or visit Cell Sciences
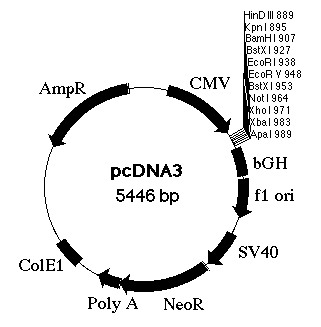
An expression vector is a plasmid that contains the properly positioned transcriptional and translational control sequences for protein expression. In order to produce bacterial proteins, a cloned structural gene must be inserted into the expression vector. With the use of a relaxed control plasmid and an efficient promoter (a type of transcriptional control element), the production of a protein of interest may reach a high level of the host’s total cellular protein.
Expression vectors may also be used to isolate specific cDNAs. We can identify specific eukaryotic cDNAs by looking for their gene products in bacteria after cloning the cDNA into appropriately constructed plasmids or phages (expression vectors). cDNAs are inserted into these vectors within regions that promote their expression in the cell (often E. coli). Often regulated bacterial promoters are used. Proteins may be expressed as fusion proteins in which amino acids from a prokaryotic protein are incorporated at one end of the eukaryotic protein. Fusion proteins are often more stable than the corresponding eukaryotic protein in bacteria and are therefore produced at higher levels.

The Gene Gun is used for the transfer of genes into living organisms. The gun uses low-pressure helium to eject cartridges of gold nano-particles into the desired object. 12 DNA or RNA-coated gold cartridges can be inserted into the holder. The Helios Gene Gun � is able to precipitate the same reaction varieties of different nucleic acids together into the subject. This enables different types of plasmids to be transferred at the same time in one experiment and thus deliver plasmids carrying various traits all in one experiment. The ability to modify the genetic makeup of cells through gene transfer can help us gain a broader understanding of gene therapy. Also, small amounts of DNA are required, thus eliminating the need for protein purification.
Visit the Bio-Rad website for more information.
Immunofluorescence staining is the use of a fluorescent antibody (i.e. antibody conjugated with a fluorochrome) for the detection and/or quantification of a specific antigen (or antibody). In the simplest immunolourescence techniques, used to detect antigen, the specimen (tissue section, smear, etc) is exposed to the conjugate (i.e. dyelinked antibody) for an appropriate time, washed free of conjugate, and examined by fluorescence microscopy; antigens homologous to the fluorescent antibodies are readily identified by regions of fluorescence in the specimen.
In the indirect immunofluorescence technique, the presence of a given antigen can be determined by first exposing the specimen to unlabeled antibodies; the specimen is then washed free of unbound antibodies, exposed to fluorescently labeled anti-Ig antibodies, washed, and examined by fluorescence microscopy. Any antigen-antibody combination on the specimen can be detected by the presence of fluorescent antibodies (which bind to unlabeled antibodies); if particular antigens are known to be present in the specimen, the test can be used to detect homologous antibodies in the serum.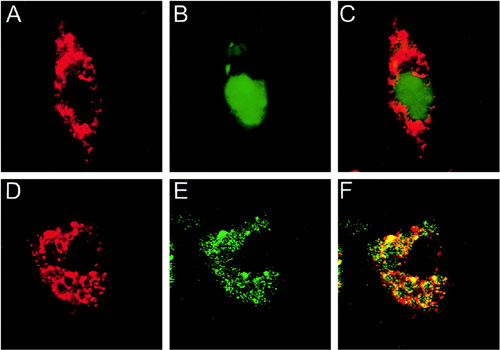
Confocal fluorescence microscopic examination to demonstrate the expression and distribution of E7 and chimeric FL-E7 proteins. 293 Db,Kb cells were transfected with pcDNA3-E7-GFP (A–C) or pcDNA3-FL-E7-GFP DNA (D–F) using Lipofectamine. Immunofluorescent staining was performed as described in “Materials and Methods.” For the detection of GFP protein, green fluorescence was noted (B and E). For the detection of endogenous calnexin protein, red fluorescence was observed (A and D). Controls omitting primary antibodies did not demonstrate specific red fluorescence (data not shown). Colocalization of GFP and calnexin was demonstrated by the yellow color in the combined image (C and F). (Enhancement of DNA Vaccine Potency by Linkage of Antigen Gene to a Gene Encoding the Extracellular Domain of Fms-like Tyrosine Kinase 3-Ligand)
Intracellular cytokine staining in this lab is perfoemd using the Cytofix/Cytoperm kit according to the manufacturer’s protocol BD PharMingen. The first step in intracellular cytokine staining is treating T cells to inhibit protein export so that cytokine accumulates within the cells. Afterwards, the membrane of T cells is permebalized with detergent, followed by labeling of cytokines within the cells using fluorochrome-labeled cytokine-specific antibodies. Cells can then be sorted by flow cytometry analysis, using a cell sorter that separates and counts labeled cells by laser and photomultiplier tube detectors, as shown in this figure with staining for CD8 and intracellular interferon gamma.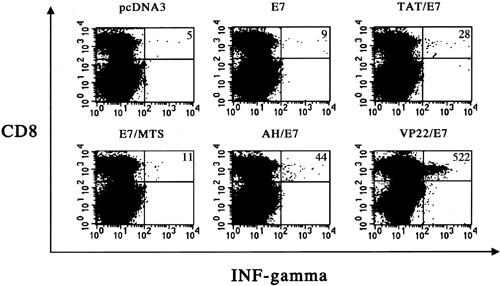
Flow cytometry analysis of IFN–secreting E7-specific CD8+ T cell precursors in mice vaccinated with various recombinant DNA vaccines. Mice were vaccinated via gene gun with 2 µg of pcDNA3, E7, TAT/E7, E7/MTS, AH/E7, or VP22/E7 DNA. One week later, mice were boosted with the same regimen as the first vaccination. Splenocytes from vaccinated mice were cultured in vitro with 1 µg/ml of E7 peptide (aa 49–57) overnight and analyzed for both CD8 and intracellular IFN- by flow cytometry analysis. Vaccination of mice with VP22/E7 DNA generated the greatest number of IFN-+ CD8+ double-positive T cells compared with the other vaccination groups. The data from intracellular cytokine staining shown here are from one representative experiment of two performed. (Improving Vaccine Potency Through Intercellular Spreading and Enhanced MHC Class I Presentation of Antigen)
Bioluminescence imaging is an non-invasive approach used to measure tumor load and distribution in mice. Murine cervical cancer models such as TC-1 tumor cell line can be modified to express luciferase. We have generated a few cell lines that express luciferase. After inoculation of mice with tumor cells expressing luciferase, D-luciferin is given to mice for activation by luciferase in the tumor cells. Seven to 8 minutes after adding luciferin, in vivo bioluminescence imaging is conducted on a cryogenically cooled IVIS Imaging System 200. The mice are placed onto the warmed stage inside a lighttight camera box with continuous isoflurane exposure. The levels of light from the bioluminescent cells are detected, integrated, and digitized. Regions of luminescence are quantified as total photon counts using acquisition and analysis software, and the values are graphed. We and others have shown that bioluminescence intensity correlates with tumor volume.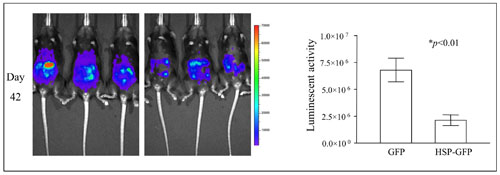
In vivo bioluminescence images of mice inoculated with luciferase-expressing tumor cells and treated with irradiated tumor cell-based vaccine. C57BL/6 mice were intraperitoneally challenged with luciferase-expressing tumor cells. Five days later, the mice were treated with irradiated tumor cells expressing Hsp70. Mice were imaged using the IVIS Imaging System Series 200. Bioluminescence signals were acquired for one minute. The figure shows luminescence images in representative mice from day 42 in the control and treated groups. Bar graphs to the right depicting the quantification of luminescent activity in the mice.
MHC: peptide tetramers consist of four peptide-bound MHC complexes adjoined to avidin or streptavidin by biotin. Flurochrome-labeled MHC:peptide tetramers are bound by receptors on T cells and can be detected and counted using flow cytometry analysis. This technique allows for the direct enumeration of antigen-specific T cells by their receptor specificity, rather than through other more indirect assays.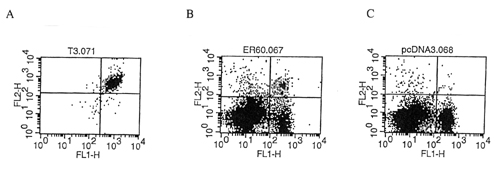
Flow cytometry analysis using double staining with CD8-specific antibody and E7 peptide-pulsed Db tetramer. X-axis represents CD8 staining an y-axis represents tetramer staining. A) E7-specific T cell clone. B) Splenocytes from mice vaccinated with an effective E7-containing DNA vaccine. C) Splenocytes from mice vaccinaated with control plasmid.
Vaccinia viruses may serve a useful viral vector since large genes can be incorporated stably into the genome, resulting in efficient replication and gene expression. Vaccinia recombination plasmids are used for placing a foreign gene into a nonessential region of the parental wild-type vaccinia virus. The plasmids contain a cloning site for insertion of the gene of interest, a selectable marker gene (eg, LacZ) or antibiotic resistance gene, and flanking portions of a nonessential vaccinia virus DNA. Co-transfection of the recombinant plasmid and a wild-type vaccinia virus into susceptible cells in culture leads to homologous recombination between the plasmid and the vaccinia genome. Selection of recombinant viruses is possible using the selectable markers found only on the recombinant plasmid. The recombinant viruses can then be purified and characterized for gene expression. Attenuated vaccinia virus (i.e. MVA, Wyeth) have also been developed for use as vaccines, are less pathogenic than wild-type virus, and are useful for human vaccination.
Western blot (or immunoblotting) is used for the detection of specific proteins. After SDS-PAGE, the gel is transferred to a nitrocellulose membrane or other solid support. Proteins are detected by treatment with specific antibodies and revealed using labeled anti-Ig antibodies.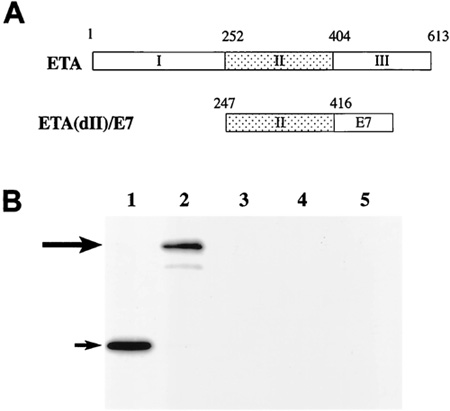
Chimeric ETA(dII)/E7 DNA construct and characterization of E7 protein expression. A, schematic diagram showing the constructs of full-length ETA and the chimeric ETA(dII)/E7 gene. The DNA fragment encoding ETA(dII) (aa 247–416) is depicted in the . The fragment encoding HPV-16 E7 (aa 1–96) is depicted in the . B, Western blot analysis to characterize the expression of E7/GFP protein in cells transfected with E7/GFP or ETA(dII)/E7/GFP DNA. Lane 1, lysates from cells transfected with E7/GFP DNA; Lane 2, lysates from cells transfected with ETA(dII)/E7/GFP DNA; Lane 3, concentrated culture medium from cells transfected with E7/GFP DNA; Lane 4, concentrated culture medium from cells transfected with ETA(dII)/E7/GFP DNA; Lane 5, lysates from nontransfected 293 Db, Kb cells as a negative control. Note: lysates from E7/GFP DNA-transfected 293 Db, Kb cells revealed a protein band with a size of approximately Mr 30,000 corresponding to E7/GFP protein in Lane 1, as indicated by the short arrow. Meanwhile, lysates from ETA(dII)/E7/GFP DNA-transfected 293 Db, Kb cells generated a protein band with a size of approximately Mr 56,000 corresponding to ETA(dII)/E7/GFP protein in Lane 2, as indicated by the long arrow. E7/GFP DNA-transfected cells exhibited levels of protein expression comparable with that of ETA(dII)/E7/GFP DNA-transfected cells.(Cancer Immunotherapy Using a DNA Vaccine Encoding the Translocation Domain of a Bacterial Toxin Linked to a Tumor Antigen)
