In eukaryotes, genomic DNA is associated with specific nuclear proteins, histones, and organized into defined structures called chromatin. Chromatin then further condenses into chromosomes. In addition to the histones involved in packaging DNA, chromosomes are also associated with many non-histone proteins required for gene expression, DNA replication, and DNA damage repair.
A class of these non-histone nuclear proteins with ATP-dependent chromatin remodeling function has been recently identified as frequently mutated genes in human cancers. Gynecologic malignancies associated with endometriosis, including clear cell carcinoma and endometrioid carcinoma, were shown by our group and others to harbor frequent inactivating mutations in ARID1A, a member of the SWI/SNF chromatin remodeling protein 1,2. Another class of non-histone nuclear proteins that consists of enzymes that catalyze covalent modification of histone N-terminal tails was also found to be mutated in a subset of cancers. Genetic alterations of these epigenetic modifiers can re-program the epigenome of cancer cells to promote tumorigenesis.
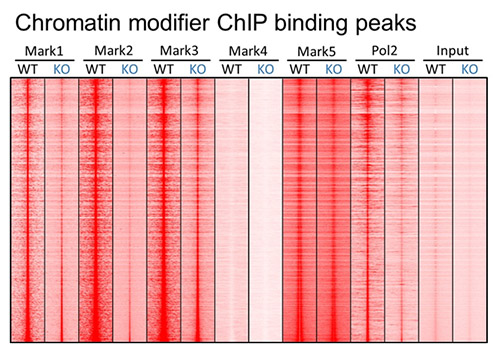
To understand the role of epigenomic regulation and alterations in cancer initiation and progression, we have employed a system biology approach by employing multiple genome-wide next generation sequencing (NGS) methods, including ChIP-seq, Faire-seq, ATAC-seq, and RNA-seq, to comprehensively elucidate the evolution of the chromatin landscape and transcriptome regulated by chromatin modifiers during tumor initiation and progression.
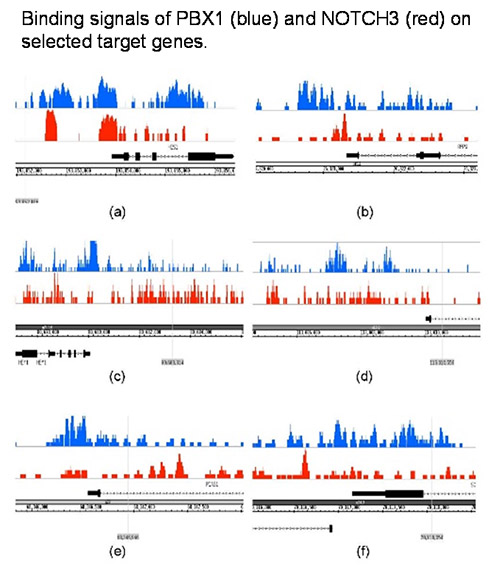
We are also exploring the functional roles of the NOTCH3 and PBX1 signaling pathways in ovarian cancer using similar approaches to understand the transcriptional networks regulated by these two signaling pathways 3,4.
References
- Jones S, Wang TL, Shih Ie M, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. Oct 8 2010;330(6001):228-231.
- Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. New Eng J Med. Oct 14 2010;363(16):1532-1543.
- Thiaville MM, Stoeck A, Chen L, et al. Identification of PBX1 target genes in cancer cells by global mapping of PBX1 binding sites. PLoS One. 2012;7(5):e36054.
- Chen X, Jung JG, Shajahan-Haq AN, et al. ChIP-BIT: Bayesian inference of target genes using a novel joint probabilistic model of ChIP-seq profiles. Nucleic Acids Res. Apr 20 2016;44(7):e65.
