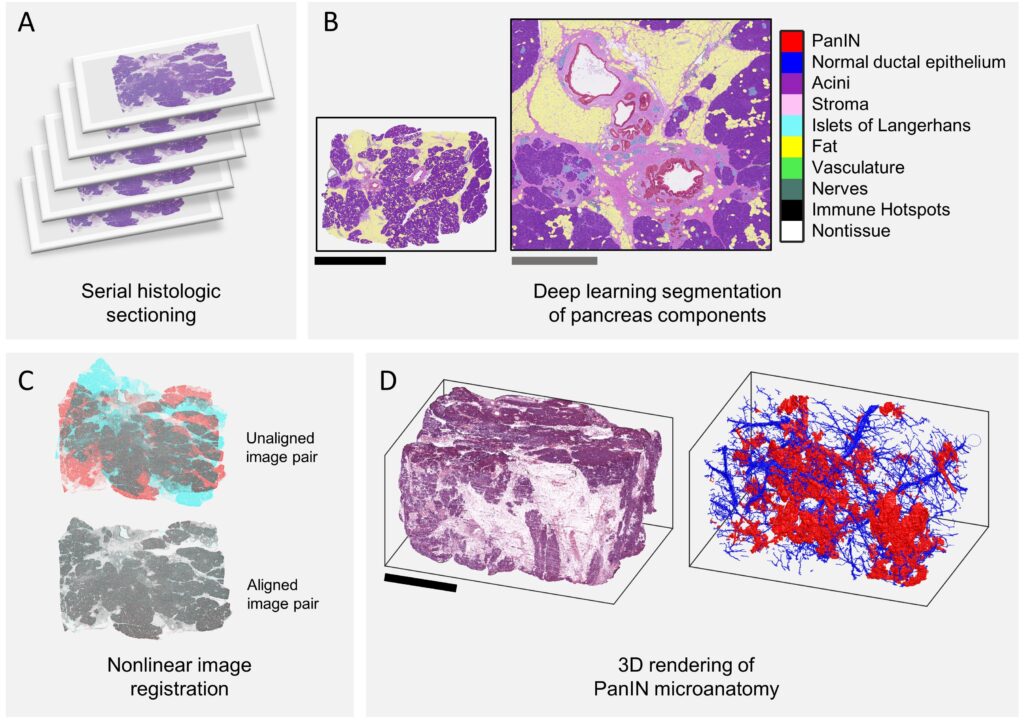
CODA is a technique for quantification of tissue microanatomy using serial histological slides, and was first described in Kiemen et al, Nature Methods (2022). To create up to cm3-sized maps of tissues at cellular resolution, CODA utilizes:
(1) nonlinear image registration
(2) deep learning semantic segmentation of microanatomical components
(3) detection of nuclear coordinates


Instructions for using CODA
The codes underlying CODA registration, tissue multilabelling, and nuclear detection are available on GitHub.
A step-by-step guide for applying CODA is available for download here.
