The Halushka Laboratory has worked with Drs. Alexander Baras, Toby Cornish and Matthew McCall to create useful software to characterize expression of microRNAs and proteins. Here are links to the tools:
Find much of our code at GitHub.
- HPAStainR – HPAStainR is a Bioconductor R Package and Shiny App used to query the scored staining patterns in the Human Protein Atlas with multiple proteins/genes of interest.
- Cell specific microRNA expression tracks at the UCSC Genome Browser
- Please follow this link and connect to the Human cellular microRNAome barChart track. And stay tuned for even more data from additional sources.
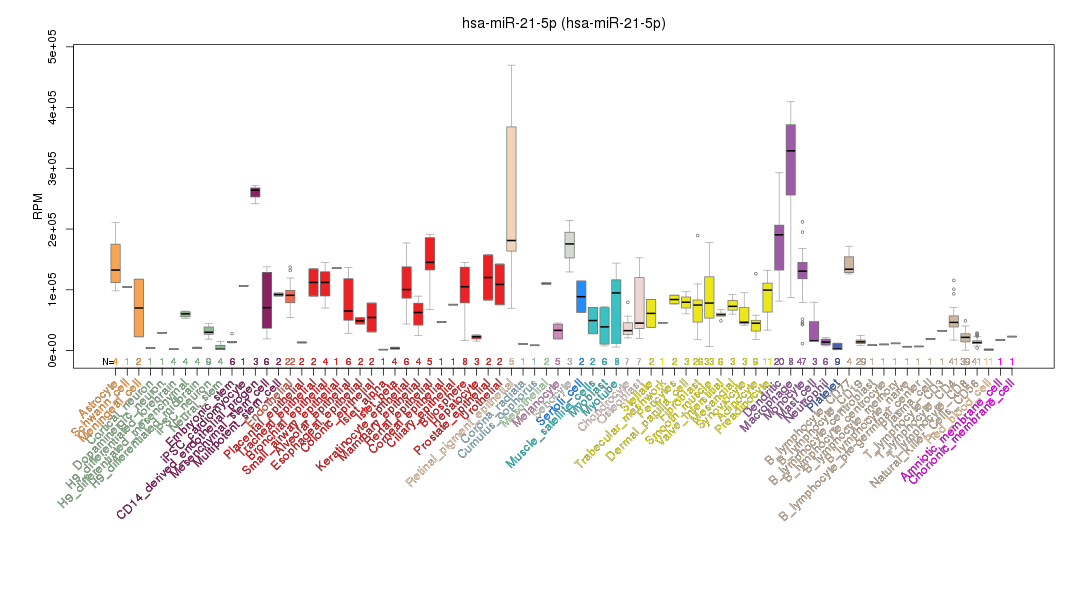
- Cellular microRNAome BioConductor package for R
- You can find it here.
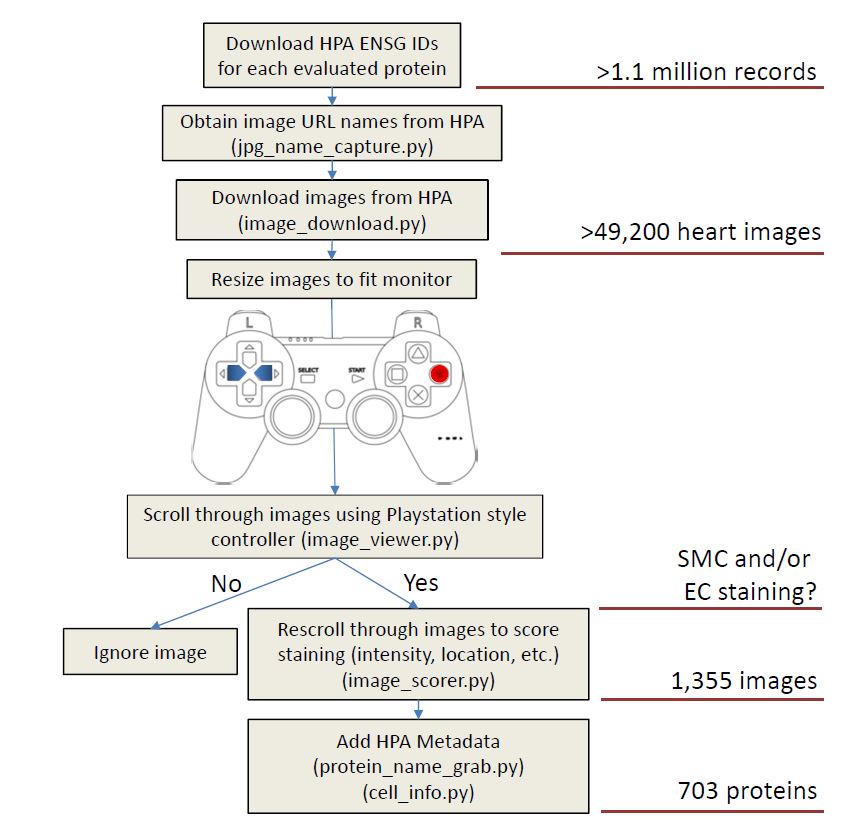
- HPASubC – HPASubC allows for rapid step-through analysis of the Human Protein Atlas image bank of protein expression. The tool can be found here: https://github.com/cornish/HPASubC.
- miRge 3.0 – This is our third generation micorRNA alignment program with scores of new and improved features. The tool can be found here. miRge 1.0 can be found here and miRge 2.0 can be found here.

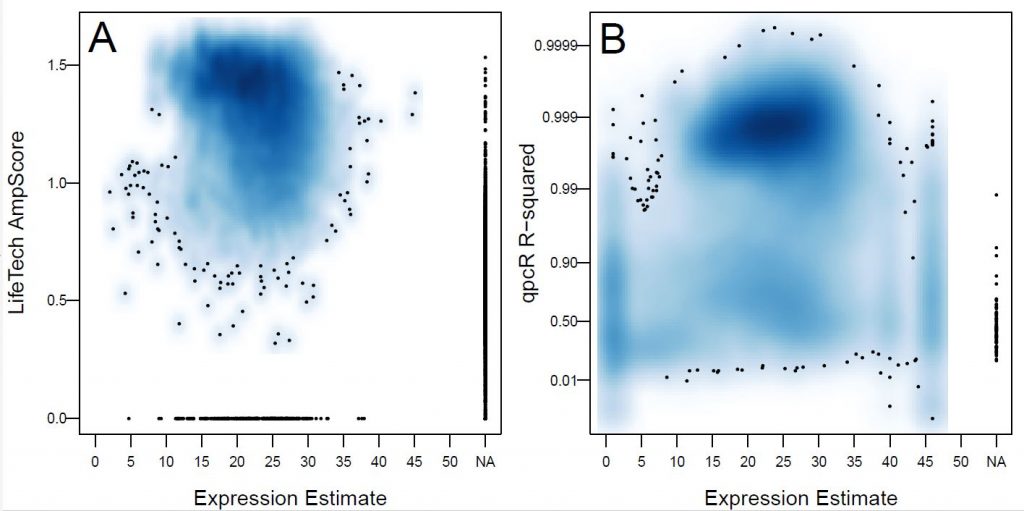
- miRComp – miRComp is a tool to assess the performance of methods that estimate expression from amplification curves derived specifically for the Life Technologies OpenArray platform. The tool can be found here: http://bioconductor.org/packages/release/bioc/html/miRcomp.html.