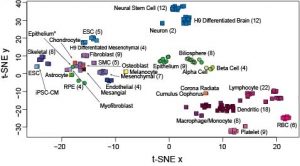
The Halushka laboratory is engaged in a number of exciting scientific endeavors. These are current lab projects:
- Contamination of the GTEx Dataset. We have recently demonstrated, using the GTEx dataset, that samples prepped and sequenced at the same time contaminate each other in small, but meaningful ways. Read more here.
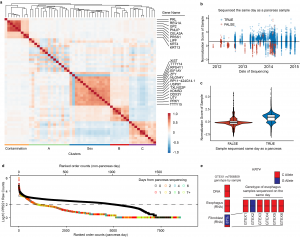
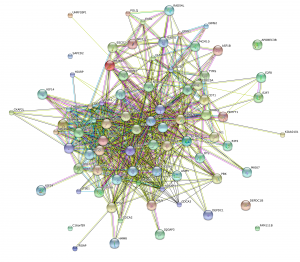
- Proteogenomic analysis of skeletal muscle. We have evaluated single skeletal muscle myocytes for their expression patterns based on gene and protein information.

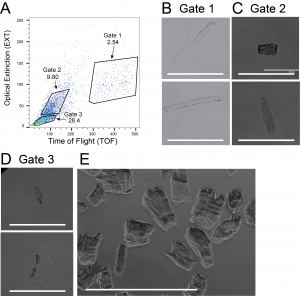
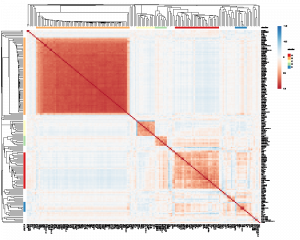
- Tissue expression deconvolution. We have been collaborating with Matthew McCall to investigate GTEx tissues to understand tissue expression composition and how to deconvolute tissue signal into its intrinsic cell parts. This work resulted in the recent AJHG manuscript entitled "Complex Sources of Variation in Tissue Expression Data: Analysis of the GTEx Lung Transcriptome".
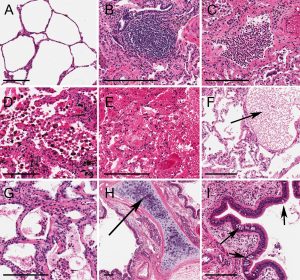
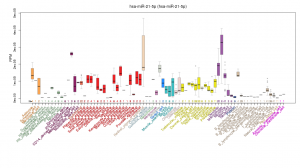
- xMD-miRNA-seq and the microRNAome. We have begun classifying the miRNA expression pattern of every human cell type. This work began in cultured cells resulting in a publication in Genome Research and a useful track on the UCSC Genome Browser. We are now exploring obtaining near in vivo miRNA cell expression data using a tool we developed called xMD-miRNA-seq.